Gro
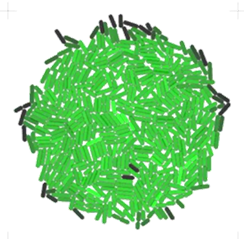
Gro is a software platform for creating and simulating agent-based models (ABMs) of bacterial colonies, widely used in both research and education. Originally developed at the Klavins Lab, University of Washington [Jang et al., 2012], Gro is known for its user-friendly desktop interface, real-time visualisation, and a simplified functional language for experiment specification.
Extended version (2017)
An extended version was later developed by our group, introducing a protein-action–based specification layer and a heuristic physics engine that significantly accelerates simulations.
Publication: Martín Gutiérrez et al., 2017
Code and Windows executable: https://github.com/liaupm/GRO-LIA
Gro 3.0
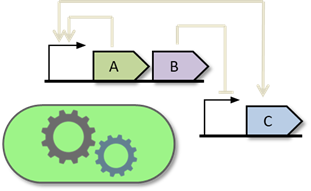
We are currently developing gro 3.0, which features an enhanced graphical interface and a novel specification layer based on biocircuit and experiment graphs. This new paradigm enables immediate, programming-free yet flexible simulations of gene expression, metabolism, ecological interactions, quorum sensing, plasmid dynamics, mutation and gene editing, bacterial conjugation, and other forms of horizontal gene transfer.
More information (doctoral thesis): https://tesis.biblioteca.upm.es/tesis/10986
Code, Windows executable, companion software, documentation and example experiment files: https://github.com/liaupm/gro-v3
YouTube channel: GRO 3 – LIA-UPM